A little while ago I came across a really cool way of trying to make sense of the impact of hyperparameters on a model. The idea is from this paper, and it is basically treating every hyperparameter as a predictor and the performance of a model on some task as the target variable. Subsequently they fit a linear model and perform an ANOVA ablation over all parameters to estimate the impact of each individual parameter for model performance. However, their code is written in r and furthermore I am not entirely sure whether the ANOVA analysis is appropriate for that study, given that the ANOVA estimate varies a lot with the order of the parameters being fitted.
Long story short, I decided to start another side-project and roll my own :). The following is based on the paper "The Dominance Analysis Approach for Comparing Predictors in Multiple Regression" (The paper is paywalled, I strongly recommend you don't try to find it on Sci-Hub in order to access this paper and many many other paywalled research papers for free). In future, I'll add some alternative estimation techniques like a causal interpretation model and perhaps other things.
The code for the whole thing is on Github, and its currently in a very experimental (though working) state. Anyhow, proceed at your own risk :). Note, the currently best way to install it is to clone the repository and then run pip install -e . inside the repository. That will install all dependencies alongside paralysis itself.
The most important thing is a file that lists all model hyperparameters and their values, together with the final performance of the model. An example file is here, which is based on running a Linear BoW SVM model with a few different parameterisations on the SNLI dataset. The example_data folder in the repository contains a few more examples. For running and recording experimental results I highly recommend using sacred.
SNLI is a textual entailment dataset and its setup as a text classifcation problem. We are given 2 sentences (the premise and the hypothesis) and have to decide whether the hypothesis ("People are playing at the seafront.") is definitely true given the premise ("5 men are playing frisbee on the beach."), whether there is a contradiction ("A group of women is crossing a busy road."), or whether we don't have enough information to make a decision ("Some people are doing something."). The SVM example uses 4 different hyperparameters, the C value, the composition_method (e.g. the way in which the premise and the hypothesis in SNLI are combined), the ngram range to consider and whether the counts in the BoW model should be binarised or not.
Running the parameter analysis is really simple and the repository also contains an example notebook.
from paralysis.parameter_ablation import ParameterAnalyser
pa = ParameterAnalyser(data='/some/path/example_data/snli_svm.json', label_name='result') # label_name specifies the name of the target variable in the results file
pa.fit_ols() # This fits the linear model on the given observations, its as simple as that!
# Now we can print an overview of which parameter is most important
print(pa.parameter_table_)
# Output
# parameter weight
# ----------------------------------
# composition_method 0.998837
# C 0.001163
# ngram_max 0.000000
# binarise 0.000000
Instead of just staring at some numbers, paralysis also has some rudimentary plotting functions to visualise the importance of each invidiual parameter.
from paralysis import visualisation
visualisation.create_plot(pa.parameter_table_)
Which results in the plot below:
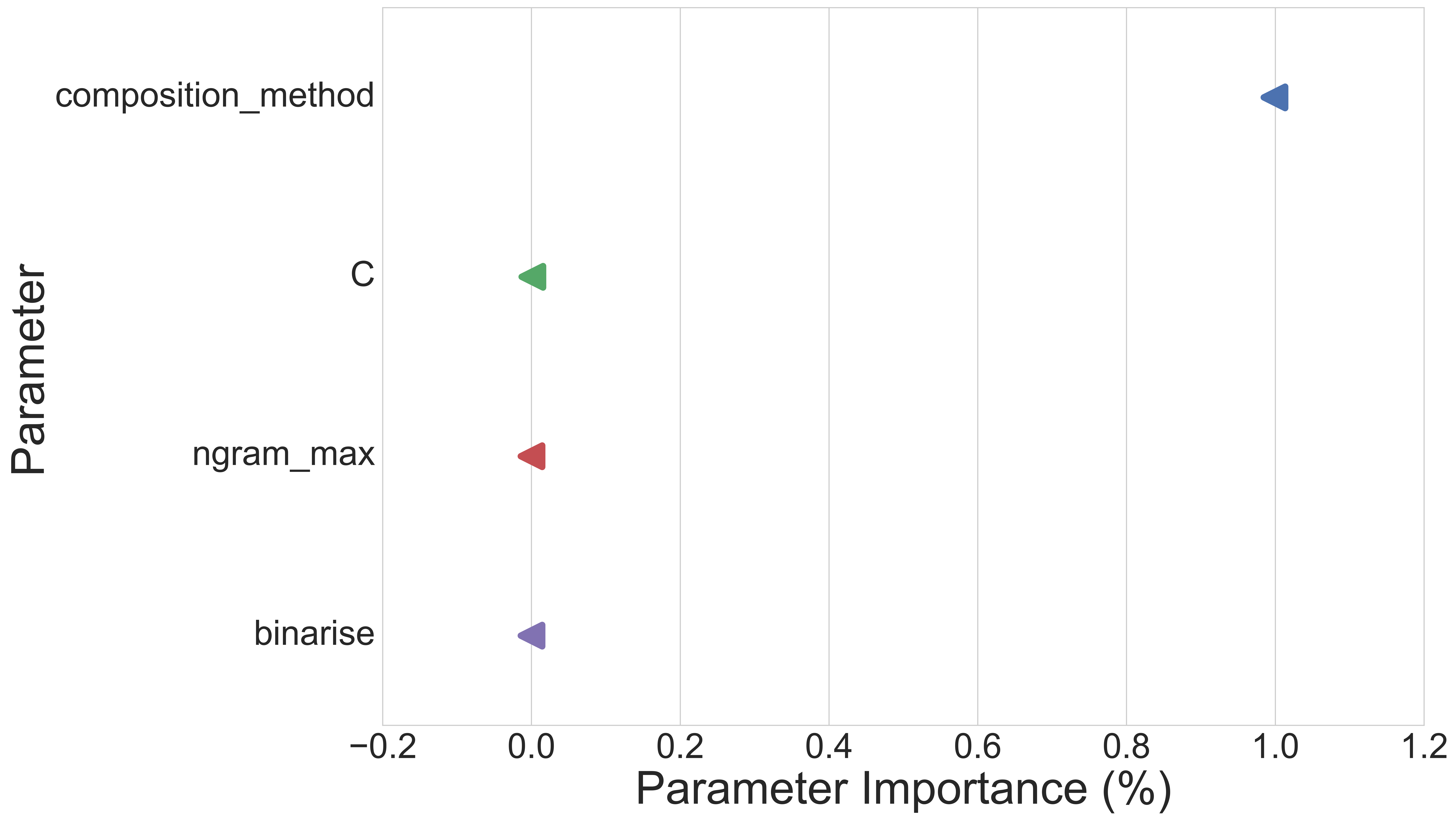
Now looking at both, the table and the plot, the weakness of the dominance analysis method that I've used becomes quite apparent. While it does correctly identify the most important parameter (and you can validate this by looking at the data), it somewhat overestimates its actual importance. Nonetheless, it can provide some first insight on a new model on which parameters are probably worth optimising, and for which others some default value suffices. One feature not listed in the example (because its not really tested yet), is to not just consider each parameter in isolation but to study their interactions. This can be done by creating the ParameterAnalyser object like pa = ParameterAnalyser(data='/some/path/example_data/snli_svm.json', label_name='result', feature_interaction_order=2'), which considers all pairs of parameters in addition to every parameter individually.